Eukaryotic genomes are organized into the 3D genome structure that is known to participate in various nuclear events including transcriptional regulation, DNA replication, and repair. Also, the disorganization of the genome structure is connected to human diseases, including various cancers and developmental syndromes. Therefore, understanding of the 3D genome organization is clearly important for the basic and medical research fields, but genome-organizing mechanisms and functions of the genome structures in distinct nuclear events are largely unclear.

Three-dimensional genome organization and functions in nuclear events
Condensin and cohesin are loaded onto genomic regions by their recruiting factors. They mediate gene contacts and the formation of topological domains. The 3D genome structures organized by condensin and cohesin participate in chromosomal dynamics, transcriptional regulation, DNA repair, and other nuclear events.
Protein complexes consisting of the structural maintenance of chromosomes (SMC) proteins are essential for the faithful segregation of chromosomes. Two of the best-studied SMC complexes are condensin and cohesin, which are required for mitotic chromosome assembly and for holding sister chromatids together, respectively. Besides the textbook paradigms, recent studies indicate that these SMC complexes are involved in genomic contacts including enhancer-promoter interactions and domain formation. However, how condensin and cohesin establish the 3D genome structure and how they contribute to nuclear events remains largely unclear. Therefore, our ongoing research projects are to determine how they mediate the 3D genome organization. We have applied genomic, genetic, cell biological, and biochemical approaches to human cells and the fission yeast model system.
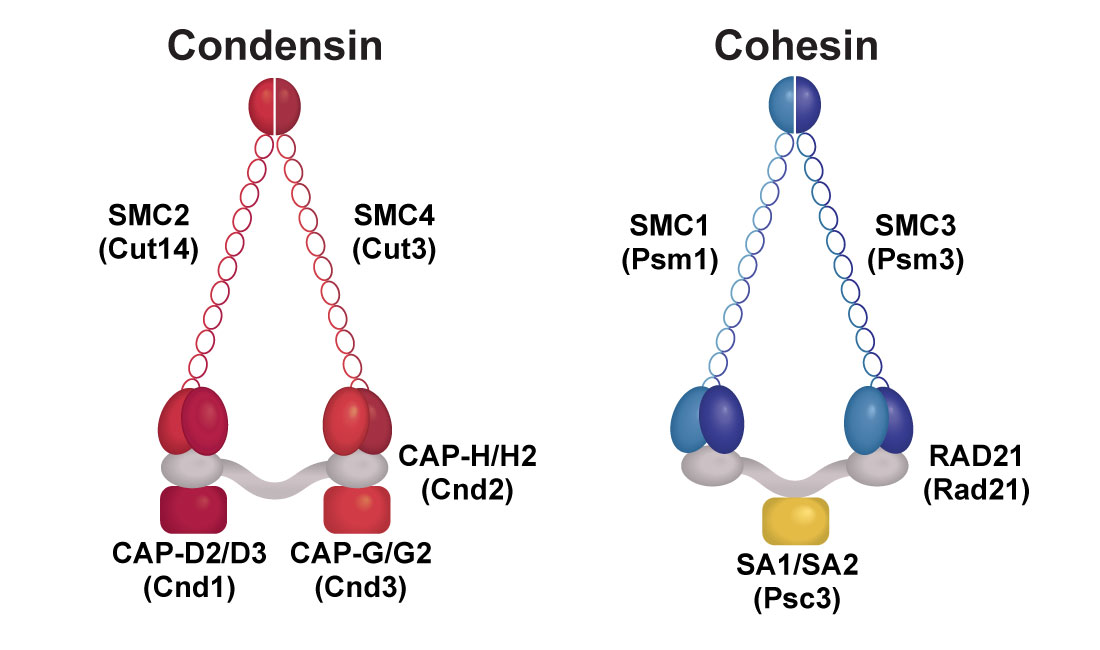
Condensin and cohesin complexes
Human condensin components are indicated above fission yeast components (in parentheses). Condensin I and II complexes exist in humans; Condensin I consists of SMC2, SMC4, CAP-D2, CAP-G and CAP-H, while condesin II contains CAP-D3, CAP-G2 and CAP-H2 in addition to the common subunits, SMC2 and SMC4.
Cellular Senescence in Humans
Cellular senescence was defined by Dr. Leonard Hayflick, as human fibroblasts cannot proliferate infinitely but reach “the Hayflick limit” and senesce. These days, cellular senescence is known as a state of very stable cell-cycle arrest caused by exhausting the capacity of DNA replication or by various cellular stresses such as DNA damage and oncogene expression. Our ongoing research focuses on the two types of senescence that are oncogene-induced senescence (OIS), which is induced by oncogene expression, and replicative senescence (RS), which is induced by telomere shortening.

SA-ß-gal and DAPI staining of senescent cells
SA-ß-gal staining was performed to detect senescent cells (left). IMR90 human lung fibroblasts were employed for this analysis. IMR90 growing, RS and OIS cells were also subjected to DAPI staining (right).
Cellular senescence is an important tumor suppression mechanism that limits the propagation of cells subjected to insults such as oncogene activation. Senescent processes are known to drastically alter expression of many genes, especially genes encoding the Senescence-Associated Secretory Phenotype (SASP) factors. The SASP factors include interleukins, chemokines, growth factors, and matrix metalloproteinases. In addition, p53 target genes are up-regulated in senescent cells. SASP, p53 target and other genes up-regulated and involved in senescent processes are collectively referred to as senescence genes.
Senescent processes also entail significant alterations of the global 3D genome structure, such as the formation of Senescence-Associated Heterochromatic Foci (SAHF) and the distension of centromeric satellites. Therefore, it appears that the 3D genome reorganization occurs upon senescence, but how the specific genome structure is established in senescent cells and contributes to the gene regulation that is characteristic of the senescent state remains unclear.
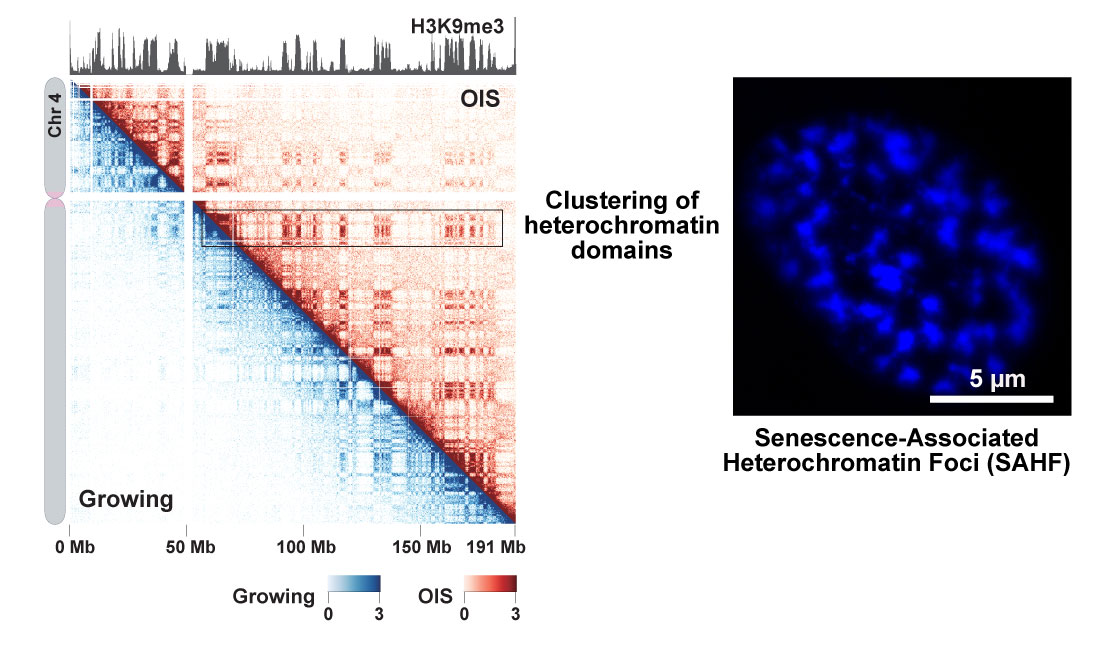
Global genomic reorganization upon cellular senescence
In situ Hi-C experiments were performed with IMR90 growing and oncogene-induced senescence (OIS) cells. Contact maps for chromosome 4 in OIS (top right) and growing cells (bottom left) are shown at 200 kb resolution. Global H3K9me3 patterns are similar between OIS and growing cells and shown on top. The in situ Hi-C data show that the clustering of heterochromatin domains is stronger in OIS cells than in proliferating cells. This means that heterochromatin aggregation visualized by DAPI staining (SAHF; right panel) can also be detected by in situ Hi-C genomics approach.
Our recent study shows how the key genome-organizing machinery, condensin, participates in the establishment and maintenance of the senescent state (Yokoyama et al. Cell Cycle 2015; Iwasaki et al. Nature Communications 2019). In addition, condensin maintains the 3D genome structure in senescent cells. Moreover, condensin is required for full activation of senescence genes including p53 targets and SASP genes. Our ongoing projects are to elucidate how condensin concomitantly regulates senescence genes and the 3D genome structure in senescent cells. Ultimately, we aim to understand how the disruption of condensin-mediated 3D genome organization contributes to oncogenic processes.
Projects Related to Cellular Senescence
- Elucidate how condensin regulates p53 targets and SASP genes in human senescent cells.
- Elucidate how condensin regulates the 3D genome structure in human senescent cells.
- Determine how condensin contributes to both Oncogene-Induced Senescence (OIS) and Replicative Senescence (RS).
3D Genome Mechanisms in the Fission Yeast Model Organism
Condensin and cohesin are the important molecular machines that establish the 3D genome architecture. Using a chromatin interaction analysis by paired-end tag sequencing (ChIA-PET) genomic approach, we show that cohesin and condensin often localize at the same loci, but they tend to mediate local and long-range gene contacts, respectively, and generate small and larger chromatin domains (Kim et al. Nature Genet 2016).
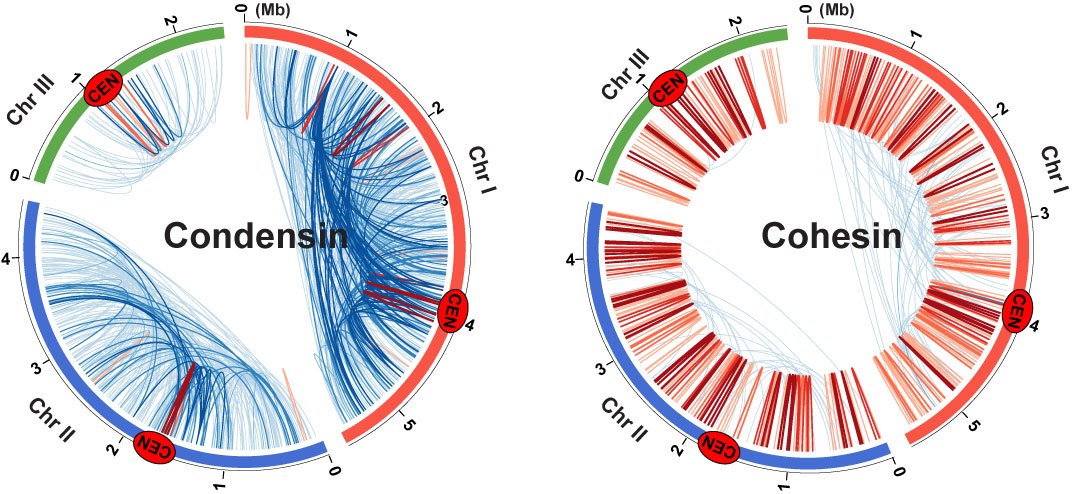
Genomic contacts mediated by condensin and cohesin
We employed the ChIA-PET genomic method to map condensin (left) and cohesin (right)-mediated gene contacts throughout the fission yeast genome. Blue and red lines represent gene contacts between two gene loci positioned more than and less than 100 kb away, respectively. Cen indicates centromeres.
We also performed genome-wide chromosome conformation capture (in situ Hi-C) and found that condensin-organized chromatin domains are promoted during mitosis and serve as chromosomal compaction units (Tanizawa et al. Nature Structural & Molecular Biology 2017). Please note that how condensin mediates mitotic chromosomal assembly is a long-standing question in the chromosome biology field. We will determine the condensin- and cohesin-mediated genome-organizing mechanisms, details of which were discussed in my recent review (Noma, Annual Review of Genetics 2017).
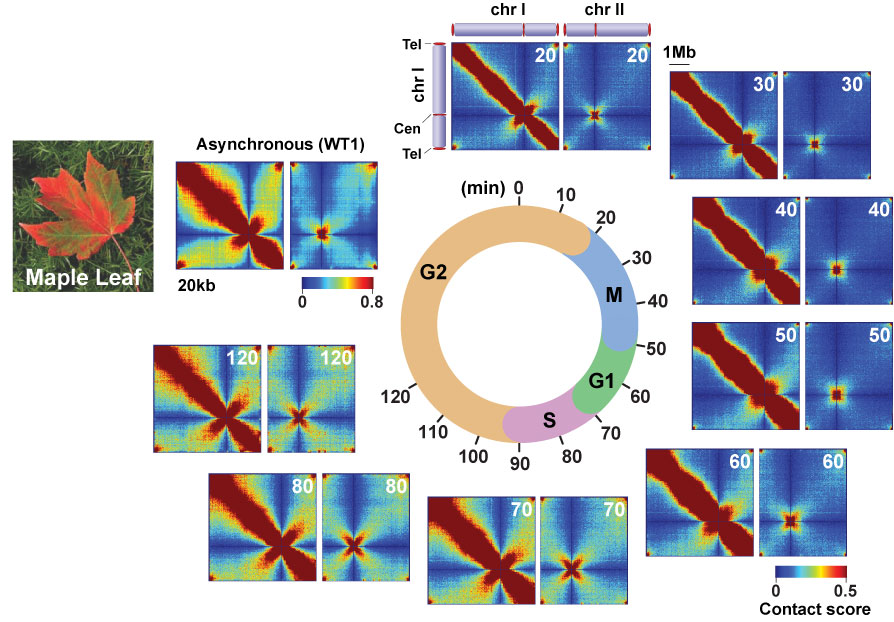
Intra- (Chr1) and inter-chromosomal (chr1 and chr2) contact maps during the cell cycle
Cells harvested at 10-minute intervals were subjected to in situ Hi-C experiments, and eight contact maps were generated throughout the cell cycle. The beautiful pattern showing genomic contacts mimics a pattern of maple leaf changing its color during autumn.
We recently found that condensin directly or indirectly interacts with a general transcription factor, TATA-box binding protein (TBP), and transcription factors (Iwasaki et al. Mol Cell 2015; Kim et al. Nature Genet 2016), but how those transcription-associated factors mediate condensin-directed genome organizations remains unclear. We will elucidate how these transcription-associated factors and condensin cooperate to establish the 3D genome structure. Our new concept is that condensin is the molecular link that bridges 3D genome organization and transcriptional regulation. More importantly, our data have demonstrated that the fission yeast mechanisms are conserved in human cells.

A model for condensin-mediated gene contacts and topological domain formation
Condensin molecules (red rings) are loaded onto genes (green boxes) and domain borders (blue boxes) via TATA box-binding protein (TBP) and transcription factors (top panel). Once they are recruited to genomic regions, they mediate gene contacts (dotted lines in middle panel), resulting in the formation of topological domains (orange sphere in bottom panel).
Projects Related to 3D Genome Mechanisms
- Elucidate how condensin mediates 3D genome organization via its interactions with TBP and transcription factors in fission yeast cells.
- Elucidate how the condensin-mediated 3D genome organization participates in transcriptional regulation.
- Elucidate how the 3D genome mechanisms characterized using the fission yeast system function in human cells.
